procedural space cannot be realized physically. there is no wear, no concept of distance. structures reside on hierarchically organized planes of existence. they can only influence other structures on the same plane or if they follow strict rules for causal interaction that must be established explicitly.
for every structure, there is a platonic ideal to which it accords. the generation of new structures is immediate. remembered by others, they exist indefinitely. without any memory of them, they vanish. existence becomes at the same time a question of relevance and of influence.
moving a structure effectively means only moving the memory others have of it. as a consequence, there is neither friction nor inertia. kinetic energy in procedural space is essentially free.
spread
#!/usr/bin/env python3
import math
def g(x: int) -> float:
if x == 0:
return 0.
return 1. / (2. ** math.ceil(math.log(x + 1., 2.)))
def h(x: int) -> int:
if x == 0:
return 0
return (2 ** math.ceil(math.log(x + 1, 2))) - x - 1
def spread(x: int) -> float:
assert x >= 0
if x == 0:
return 0.
rec_x = h(x - 1)
return spread(rec_x) + g(x)
def main():
x = 0
while True:
y = spread(x)
print(y)
x += 1
if __name__ == "__main__":
main()a tail recursion generates output values between 0. and 1. with continual integer increments starting from zero, the function generates a uniform distribution of return values.
the argument integer x is passed on to the main function spread. if x is zero, the recursion terminates with 0. if not, the greatest power of two below x is determined. the function spread is called recursively with the result as an argument. the return value from the recursive call is added to the inverse of the previous result and the function finally returns the sum of both.
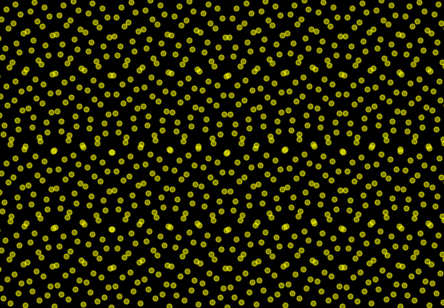
plotting the result of the function along its input integers creates a pseudo-random carpet that strikes an interesting balance between repetition and variance.
divide
#!/usr/bin/env python3
import itertools
from typing import Tuple, Generator
POINT = Tuple[float, ...]
CUBE = Tuple[POINT, POINT]
def _divide(corners: CUBE, center: POINT) -> Tuple[CUBE, ...]:
return tuple((_x, center) for _x in itertools.product(*zip(*corners)))
def _center(corners: CUBE) -> POINT:
point_a, point_b = corners
return tuple((_a + _b) / 2. for _a, _b in zip(point_a, point_b))
def uniform_areal_segmentation(dimensionality: int) -> Generator[Tuple[CUBE, POINT], None, None]:
corners = tuple(0. for _ in range(dimensionality)), tuple(1. for _ in range(dimensionality))
spaces = [corners]
while True:
_spaces_new = []
while 0 < len(spaces):
_each_cube = spaces.pop()
center = _center(_each_cube)
_segments = _divide(_each_cube, center)
_spaces_new.extend(_segments)
yield _each_cube, center
spaces = _spaces_new
def main():
dimensions = 2
generator_segmentation = uniform_areal_segmentation(dimensions)
while True:
_, _point = next(generator_segmentation)
print(_point)
if __name__ == "__main__":
main()the iterative process generates a vector inside a unit cube of arbitrary dimensions. the vectors are distributed such that the probability of reaching any point within the cube approaches an equilibrium as iterations approximate infinity.
the generator is initialized with a top-level queue that holds two corners that define a unit cube in n dimensions and an empty low-level queue. each of the last two corners in the top-level queue together with the center between them defines the two corners of 2 ** n new smaller cubes. the original corners are removed, the corners of the smaller cubes are added to the low-level queue, and the generator yields their center.
if the top-level queue is empty, it is replaced by the low-level queue which is then cleared.
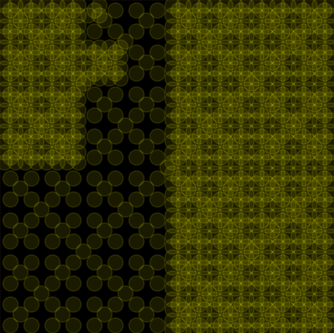
overlapping data points in two-dimensional space form a geometric pattern. the uniformly distributed points afford themselves to the unbiased sampling of a volume of arbitrary dimensions (e.g. in an optimizer for nondifferentiable functions).
enhance
#!/usr/bin/env python3
import random
from typing import Sequence, Generator, Tuple
def my_range(no_samples: int,
start: float = 0., end: float = 1.,
start_point: bool = True, end_point: bool = True) -> Generator[float, None, None]:
assert 1 < no_samples
float_not_start = float(not start_point)
step_size = (end - start) / (no_samples - float(end_point) + float_not_start)
start_value = start + float_not_start * step_size
for _ in range(no_samples):
yield min(end, max(start, start_value))
start_value += step_size
return
def single_sample_uniform(no_samples: int, mean: float,
include_borders: bool = True) -> Tuple[float, ...]:
assert no_samples >= 1
assert 0. < mean < 1.
if no_samples == 1:
return mean,
sample_ratio_right = no_samples * mean
sample_ratio_left = no_samples - sample_ratio_right
no_samples_right = round(sample_ratio_right)
no_samples_left = round(sample_ratio_left)
if no_samples_right < 1:
no_samples_right += 1
no_samples_left -= 1
elif no_samples_left < 1:
no_samples_right -= 1
no_samples_left += 1
# right biased
if .5 < mean:
if no_samples_right == 1:
samples_right = (1.,) if include_borders else (mean + 1. / 2.,)
else:
samples_right = tuple(
my_range(
no_samples_right,
start_point=False, end_point=include_borders,
start=mean, end=1.
)
)
mean_left = mean - sum(_s - mean for _s in samples_right) / no_samples_left
if no_samples_left == 1:
samples_left = max(0., mean_left),
else:
radius_left = min(mean_left, mean - mean_left)
samples_left = tuple(my_range(
no_samples_left,
start_point=include_borders, end_point=include_borders,
start=max(0., mean_left - radius_left),
end=min(mean_left + radius_left, mean),
))
# left biased
else:
if no_samples_left == 1:
samples_left = 0.,
else:
samples_left = tuple(
my_range(
no_samples_left,
start_point=include_borders, end_point=False,
start=0, end=mean
)
)
mean_right = mean + sum(mean - _s for _s in samples_left) / no_samples_right
if no_samples_right == 1:
samples_right = min(1., mean_right),
else:
radius_right = min(mean_right - mean, 1. - mean_right)
samples_right = tuple(my_range(
no_samples_right,
start_point=include_borders, end_point=include_borders,
start=max(mean, mean_right - radius_right),
end=min(1., mean_right + radius_right),
))
return samples_left + samples_right
def multi_sample_uniform(no_samples: int,
means: Sequence[float],
include_borders: bool = True) -> Tuple[Sequence[float], ...]:
assert all(1. >= _x >= 0. for _x in means)
unzipped = [
single_sample_uniform(no_samples, _m, include_borders=include_borders)
for _m in means
]
return tuple(zip(*unzipped))
def main():
dimensions = random.randint(1, 4)
target = tuple(random.random() for _ in range(dimensions))
no_samples = random.randint(4, 6)
include_borders = False
samples = multi_sample_uniform(no_samples, target, include_borders=include_borders)
print(f"{str(target):s} = ({' + '.join(str(vector) for vector in samples):s}) / {no_samples:d}")
print()
if __name__ == "__main__":
main()the process generates m vectors within a unit cube of arbitrary dimension, such that their average is equal to the target vector. the borders of the unit cube can be included or excluded.
for each dimension, the number of samples below and above the target value is determined. if the target value is below .5, then there is more space above to compensate for the samples below. therefore, more samples need to be below (in a lower distance) and fewer samples need to be above (in a greater distance) to the target value. vice versa for a target value above .5. the location of samples is determined by distributing the sum of differences between samples and target on one side among the remaining samples on the other side.
as a consequence, a sample size of one simply returns the target vector. a sample size of two returns two vectors of maximum equal distance to the target vector. sample sizes beyond two generate results that afford themselves to the procedural generation of detail.